| Gene | Ercc4 | ||||||||||||||||||||||||||||||||||||||
| Synonyms | Xpf | ||||||||||||||||||||||||||||||||||||||
| Description | excision repair cross-complementing rodent repair deficiency, complementation group 4 | ||||||||||||||||||||||||||||||||||||||
| Locus | NM_015769 UCSC: chr16:13109829-13152102(+) OLP: chr16:13109829-13152102(+) | ||||||||||||||||||||||||||||||||||||||
| DB links | 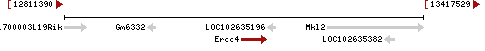 Entrez 50505 MGI 1354163 | ||||||||||||||||||||||||||||||||||||||
| Feature | protein-coding | ||||||||||||||||||||||||||||||||||||||
| GO_BP |
| ||||||||||||||||||||||||||||||||||||||
| GO_MF |
| ||||||||||||||||||||||||||||||||||||||
| GO_CC |
| ||||||||||||||||||||||||||||||||||||||
| KEGG |
| ||||||||||||||||||||||||||||||||||||||
| Ver2_FPKM |
| ||||||||||||||||||||||||||||||||||||||
| SPR_Histone_Erkek_5KaroundTSS |
| ||||||||||||||||||||||||||||||||||||||
| SPR_Histone_Erkek_2KaroundTSS |
| ||||||||||||||||||||||||||||||||||||||
| SPR_Methy_Kobayashi_5KaroundTSS |
| ||||||||||||||||||||||||||||||||||||||
| SPR_Methy_Kobayashi_2KaroundTSS |
| ||||||||||||||||||||||||||||||||||||||
| SOLiD_OocyteTo4C_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| SOLiD_Illumina_Oo_1C_Sperm_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| SOLiD_Partheno1C_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| SOLiD_Illumina_Oo_p1C_Sperm_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| SOLiD_Partheno4C_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| Illumina_Oo_2C_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| SOLiD_SC_Oo_REPLICA_FPKM_gene_guided | N/A | ||||||||||||||||||||||||||||||||||||||
| NIA60mer |
| ||||||||||||||||||||||||||||||||||||||
| MOE430 | N/A | ||||||||||||||||||||||||||||||||||||||
| Agilent44K | N/A | ||||||||||||||||||||||||||||||||||||||
| Proteome | N/A |
 16 Terms
16 Terms