DBTMEE:2010214
| Gene | Hspa5 |
| Synonyms | 78kDa
baffled
Bip
D2Wsu141e
D2Wsu17e
Grp78
Hsce70
mBiP
Sez7
XAP-1 antigen |
| Description | heat shock protein 5 |
| Locus | NM_001163434 UCSC: chr2:34627610-34632049(+) OLP: chr2:34627610-34632049(+)
NM_022310 UCSC: chr2:34627610-34632049(+) OLP: chr2:34627610-34632049(+)
|
| DB links |
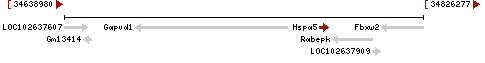
Entrez 14828
MGI 95835
Ensembl ENSMUSG00000026864
Vega OTTMUSG00000012014
|
| Feature | protein-coding |
| GO_BP |
-
 14 Terms
ER overload response (GO:0006983)
activation of signaling protein activity involved in unfolded protein response (GO:0006987)
cerebellum structural organization (GO:0021589)
cerebellar Purkinje cell layer development (GO:0021680)
positive regulation of cell migration (GO:0030335)
negative regulation of transforming growth factor beta receptor signaling pathway (GO:0030512)
positive regulation of protein ubiquitination (GO:0031398)
response to endoplasmic reticulum stress (GO:0034976)
maintenance of protein localization in endoplasmic reticulum (GO:0035437)
positive regulation of embryonic development (GO:0040019)
cellular response to glucose starvation (GO:0042149)
negative regulation of apoptotic process (GO:0043066)
proteolysis involved in cellular protein catabolic process (GO:0051603)
cellular response to interleukin-4 (GO:0071353)
|
| GO_MF |
-
 10 Terms
|
| GO_CC |
-
 16 Terms
|
| KEGG |
-
 4 Terms
|
| Ver2_FPKM |
| DBTMEE | Gene | RefSeq | Entrez | SPR_FPKM | Oo_FPKM | 1C_FPKM | 2C_FPKM | 4C_FPKM | ESC_FPKM | MEF_FPKM | p1C_FPKM | p4C_FPKM | iPSC_FPKM |
|---|
2010214
(v2.0) | Hspa5 | NM_001163434,NM_022310 | 14828
| 495.928 | 100.122 | 56.5433 | 256.682 | 210.262 | 423.223 | 479.853 | 39.9503 | 225.31 | 506.802 |
|
| SPR_Histone_Erkek_5KaroundTSS |
| DBTMEE | Gene | Entrez | H3.3_1_ChIP/Input | H3.3_1(Enrich; log2) | H3.3_2_ChIP/Input | H3.3_2(Enrich; log2) | H3K4me3_1_ChIP/Input | H3K4me3_1(Enrich; log2) | H3K4me3_2_ChIP/Input | H3K4me3_2(Enrich; log2) | H3K27me3_1_ChIP/Input | H3K27me3_1(Enrich; log2) | H3K27me3_2_ChIP/Input | H3K27me3_2(Enrich; log2) |
|---|
2010214
(v2.0) | Hspa5 | 14828
| 701/446 | 0.1393 | 473/446 | -0.0313 | 415/446 | 3.4619 | 1203/446 | 2.7559 | 11/446 | -3.1716 | 111/446 | -2.5895 |
|
| SPR_Histone_Erkek_2KaroundTSS |
| DBTMEE | Gene | Entrez | H3.3_1_ChIP/Input | H3.3_1(Enrich; log2) | H3.3_2_ChIP/Input | H3.3_2(Enrich; log2) | H3K4me3_1_ChIP/Input | H3K4me3_1(Enrich; log2) | H3K4me3_2_ChIP/Input | H3K4me3_2(Enrich; log2) | H3K27me3_1_ChIP/Input | H3K27me3_1(Enrich; log2) | H3K27me3_2_ChIP/Input | H3K27me3_2(Enrich; log2) |
|---|
2010214
(v2.0) | Hspa5 | 14828
| 360/211 | 0.2568 | 206/211 | -0.15 | 385/211 | 4.3918 | 965/211 | 3.509 | 6/211 | -2.8841 | 39/211 | -2.9869 |
|
| SPR_Methy_Kobayashi_5KaroundTSS |
| DBTMEE | Gene | Entrez | Total_mC_levels | Number_of_mCs | Averaged_Methylation_level |
|---|
2010214
(v2.0) | Hspa5 | 14828
| 63.7412 | 178 | 35.81 |
|
| SPR_Methy_Kobayashi_2KaroundTSS |
| DBTMEE | Gene | Entrez | Total_mC_levels | Number_of_mCs | Averaged_Methylation_level |
|---|
2010214
(v2.0) | Hspa5 | 14828
| 18.0865 | 130 | 13.91 |
|
| SOLiD_OocyteTo4C_FPKM_gene_guided |
N/A |
| SOLiD_Illumina_Oo_1C_Sperm_FPKM_gene_guided |
N/A |
| SOLiD_Partheno1C_FPKM_gene_guided |
N/A |
| SOLiD_Illumina_Oo_p1C_Sperm_FPKM_gene_guided |
N/A |
| SOLiD_Partheno4C_FPKM_gene_guided |
N/A |
| Illumina_Oo_2C_FPKM_gene_guided |
N/A |
| SOLiD_SC_Oo_REPLICA_FPKM_gene_guided |
N/A |
| NIA60mer |
| DBTMEE | Array_Feature_Number | Oligo_Parent_Clone | Unigene | Gene | Entrez | U_Cluster | NAP_Cluster | Gene_U/NAP | TIGR | SwissProt | EST | Oo | 1C | 2C | 4C | 8C | Morula | Blastocyst |
|---|
0010327
(v1.0) | 1979 | C0277C03-3 | Mm.918 | Hspa5 | 14828
| U042281 | NAP093911-001 | Hspa5; | TC873610,TC873602,TC873601,TC873591,TC873583,TC873583 | P20029 | U,F,2 | 3.1853 | 3.3297 | 3.7547 | 4.4173 | 3.285 | 3.5537 | 3.5473 |
0010327
(v1.0) | 5034 | G0120A06-3 | Mm.918 | Hspa5 | 14828
| U042281 | NAP093911-001 | Hspa5; | TC873610,TC873602,TC873601,TC873591,TC873583,TC873583 | P20029 | U,F,2 | 3.23 | 3.3712 | 3.7826 | 4.4397 | 3.2942 | 3.5717 | 3.5687 |
|
| MOE430 |
N/A |
| Agilent44K |
N/A |
| Proteome |
N/A |
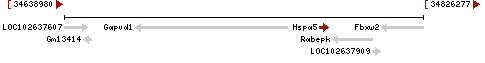
 14 Terms
14 Terms