Ultralarge-scale Whole-transcriptome Analysis of Mouse Early Embryos
[Purpose]
We develop an information analysis system to systematically examine male and female genome dynamics from fertilization to zygotic gene activation (ZGA). The system will be based on data correlation between transitions of protein localization on the chromosomes, intranuclear chromosome location, and chromosome three-dimensional conformation, from immediately after fertilization to the 8-cell stage. A model of the structural specificity of male and female chromosomes and the course leading to ZGA will be constructed using this system, and an experimental investigation will be conducted to obtain a comprehensive understanding of genome dynamics after fertilization. To facilitate a comprehensive analysis, we collect other publicly available resources, such as proteomic data, microarray profiles, and ChIP-seq data of epigenome.
[Introduction]
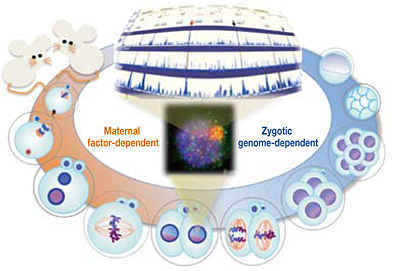
Fertilization of an oocyte by a sperm is the first developmental step to confer totipotency on the newly formed embryo. Fertilization stimulates a precisely controlled coordination of maternal and paternal genomes through a sequence of different biological processes. Deciphering the mechanisms underlying this choreography of the parental genomes in mammalian embryos has long been recognized as one of the fundamental challenges in human infertility and stem cell biology research. Large-scale genome reconstruction begins in spermatic and egg chromosomes immediately after fertilization, followed by the induction of pronuclear formation. Sperm chromosomes are highly condensed by a protein called protamine. It is believed that decondensation is induced upon fertilization, and various transcription factors and genome conformation-related proteins, including histone, start to bind to the genome, though the details are unknown. The pronuclei, containing either the male or female chromosomes, do not fuse with each other while the first chromosome segregation following DNA replication occurs. The following one or two cell cycle(s) are driven mainly by maternal factors; thus, cell division occurs in a particular "maternal-factor-dependent" manner. At the pronuclear stage, ZGA has already begun, and major zygotic genes are activated at the 2- or 4-cell stage. Meanwhile, male and female chromosomes are localized separately in respective regions without being mixed. The genome reconstruction that occurs in male and female chromosomes early after fertilization is the most basic and indispensable process necessary for the ZGA, the shift to zygotic genome-dependent cell cycles, and the development of individuals. Due to difficulties in analysis, most of the details of these processes have come only from cell biology studies.
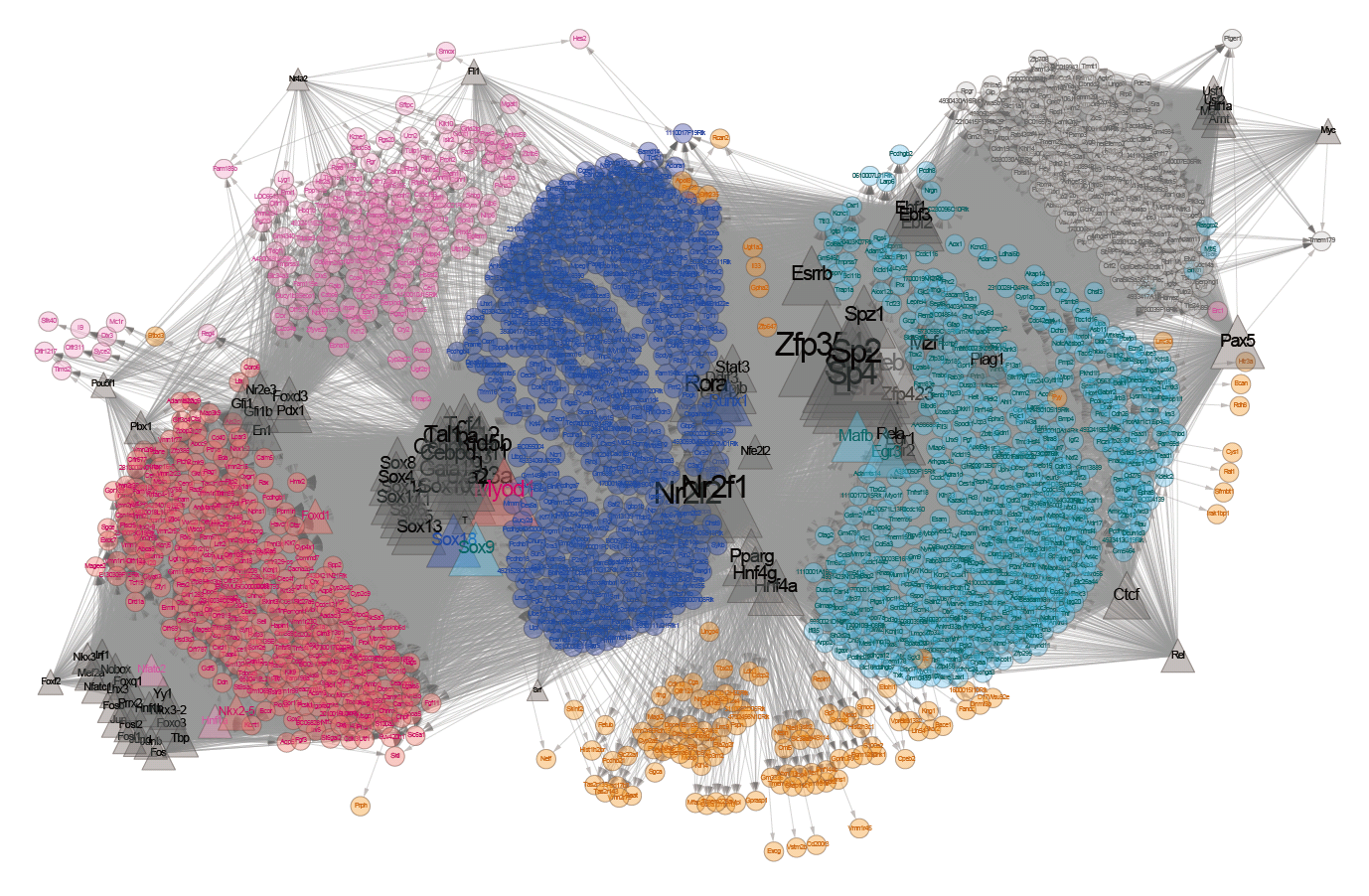
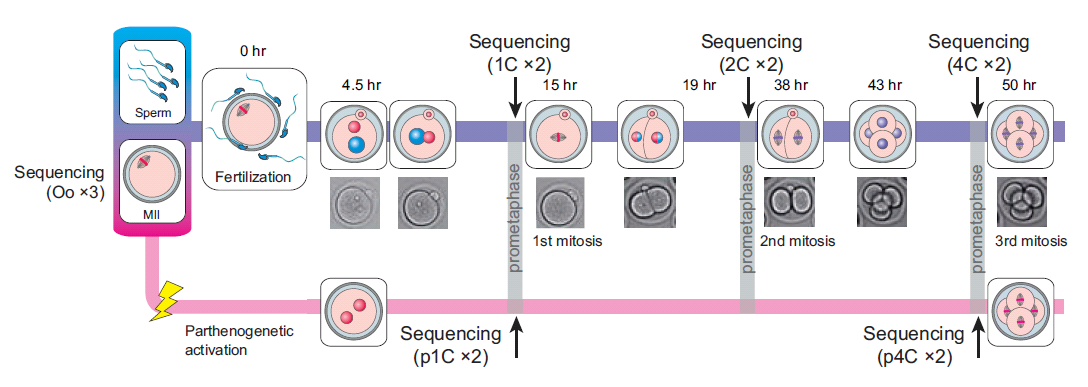
Here, we sequenced more than 1.5×105 MII oocytes to analyse the transition of male and female genome structures from fertilization through ZGA to the zygotic genome-dependent cell cycles. DBTMEE (Database of Transcriptome in Mouse Early Embryos) is a database providing the gene expression profile established by this ultralarge-scale whole-transcriptome analysis. DBTMEE also contains other public resources to systematically understand male and female genome dynamics from fertilization to ZGA. This system will be used to prepare a model of the structural specificity of male and female chromosomes and the course of events leading to ZGA. We believe that our database will help researchers explore the genetic and epigenetic characteristics of genes involved in genome activation programs and, in particular, identify potential key transcription factors (TFs) involved in transcriptional initiation in fertilized oocytes.
References1. Park SJ, Shirahige K, Ohsugi M, Nakai K,
"DBTMEE: a database of transcriptome in mouse early embryos",
Nucl. Acids Res. (2015), 43(D1):D771-776. [NAR]
2. Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K,
"Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis",
Genes Dev. 2013 Dec 15;27(24):2736-48. [PUBMED]
