DBTMEE:0003151
| Gene | Axin1 |
| Synonyms | |
| Description | axin 1 |
| Locus | NM_001159598 UCSC: chr17:26275631-26332756(+) OLP: chr17:26275631-26332756(+)
NM_009733 UCSC: chr17:26275631-26332756(+) OLP: chr17:26275631-26332756(+)
|
| DB links |
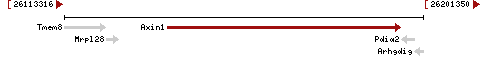
Entrez 12005
MGI 1096327
Ensembl ENSMUSG00000024182
Vega OTTMUSG00000026498
|
| Feature | protein-coding |
| GO_BP |
-
 57 Terms
protein polyubiquitination (GO:0000209)
in utero embryonic development (GO:0001701)
optic placode formation (GO:0001743)
regulation of protein phosphorylation (GO:0001932)
positive regulation of protein phosphorylation (GO:0001934)
nucleocytoplasmic transport (GO:0006913)
apoptotic process (GO:0006915)
activation of JUN kinase activity (GO:0007257)
multicellular organismal development (GO:0007275)
determination of left/right symmetry (GO:0007368)
sensory perception of sound (GO:0007605)
adult walking behavior (GO:0007628)
cell death (GO:0008219)
dorsal/ventral axis specification (GO:0009950)
dorsal/ventral pattern formation (GO:0009953)
positive regulation of peptidyl-threonine phosphorylation (GO:0010800)
Wnt receptor signaling pathway (GO:0016055)
hemoglobin metabolic process (GO:0020027)
forebrain anterior/posterior pattern specification (GO:0021797)
Wnt receptor signaling pathway involved in forebrain neuron fate commitment (GO:0021881)
protein catabolic process (GO:0030163)
negative regulation of Wnt receptor signaling pathway (GO:0030178)
positive regulation of transforming growth factor beta receptor signaling pathway (GO:0030511)
olfactory placode formation (GO:0030910)
cytoplasmic microtubule organization (GO:0031122)
positive regulation of protein ubiquitination (GO:0031398)
activation of protein kinase activity (GO:0032147)
positive regulation of proteasomal ubiquitin-dependent protein catabolic process (GO:0032436)
positive regulation of peptidyl-serine phosphorylation (GO:0033138)
erythrocyte homeostasis (GO:0034101)
negative regulation of transcription elongation from RNA polymerase II promoter (GO:0034244)
regulation of catenin import into nucleus (GO:0035412)
post-anal tail morphogenesis (GO:0036342)
termination of G-protein coupled receptor signaling pathway (GO:0038032)
positive regulation of JUN kinase activity (GO:0043507)
cellular protein complex assembly (GO:0043623)
negative regulation of fat cell differentiation (GO:0045599)
positive regulation of protein catabolic process (GO:0045732)
positive regulation of protein kinase activity (GO:0045860)
positive regulation of transcription, DNA-dependent (GO:0045893)
positive regulation of JNK cascade (GO:0046330)
embryonic eye morphogenesis (GO:0048048)
axial mesoderm development (GO:0048318)
axial mesoderm formation (GO:0048320)
neurological system process (GO:0050877)
negative regulation of protein metabolic process (GO:0051248)
protein homooligomerization (GO:0051260)
positive regulation of ubiquitin-protein ligase activity (GO:0051443)
muscle cell development (GO:0055001)
canonical Wnt receptor signaling pathway (GO:0060070)
embryonic skeletal joint morphogenesis (GO:0060272)
canonical Wnt receptor signaling pathway involved in neural plate anterior/posterior pattern formation (GO:0060823)
cellular response to organic cyclic compound (GO:0071407)
genetic imprinting (GO:0071514)
negative regulation of canonical Wnt receptor signaling pathway (GO:0090090)
Wnt receptor signaling pathway involved in somitogenesis (GO:0090244)
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process (GO:2000060)
|
| GO_MF |
-
 18 Terms
|
| GO_CC |
-
 17 Terms
|
| KEGG |
-
 5 Terms
|
| Ver2_FPKM |
see DBTMEE:2003055 (v2.0)
|
| SPR_Histone_Erkek_5KaroundTSS |
N/A |
| SPR_Histone_Erkek_2KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_5KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_2KaroundTSS |
N/A |
| SOLiD_OocyteTo4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | SPR_FPKM | Oo_FPKM | 1C_FPKM | 2C_FPKM | 4C_FPKM | MEF_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 26.8932 | 37.7909 | 39.9544 | 25.8336 | 12.943 | |
|
| SOLiD_Illumina_Oo_1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | 1C_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 28.1193 | 4.81636 | 41.2307 |
|
| SOLiD_Partheno1C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 1C_FPKM | p1C_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 27.4472 | 37.4115 | 46.021 |
|
| SOLiD_Illumina_Oo_p1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | p1C_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 28.1356 | 4.7504 | 50.4743 |
|
| SOLiD_Partheno4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | 2C_FPKM | 4C_FPKM | p4C_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 36.9915 | 24.9434 | 24.9857 |
|
| Illumina_Oo_2C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 2C_FPKM |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 17.0299 | 5.59276 |
|
| SOLiD_SC_Oo_REPLICA_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_1_1 | Oo_2_1 |
|---|
0003151
(v1.0) | Axin1 | NM_001159598,NM_009733 | 12005
| 17.749 | 11.2294 |
|
| NIA60mer |
| DBTMEE | Array_Feature_Number | Oligo_Parent_Clone | Unigene | Gene | Entrez | U_Cluster | NAP_Cluster | Gene_U/NAP | TIGR | SwissProt | EST | Oo | 1C | 2C | 4C | 8C | Morula | Blastocyst |
|---|
0003151
(v1.0) | 10976 | H3106D02-3 | Mm.23684 | Axin | 12005
| U017701 | NAP003573-001 | Axin; | TC826165 | O35625 | U,F,2 | 3.573 | 3.5501 | 3.6077 | 3.574 | 3.4901 | 3.4467 | 3.4687 |
|
| MOE430 |
| DBTMEE | Probe_ID | Unigene | Gene | RefSeq | Entrez | Oo | 1C | 2C | 8C | Blastocyst |
|---|
0003151
(v1.0) | 1426966_at | Mm.23684 | Axin1 | NM_009733.2,NM_001159598.1 | 12005
| 186.775 | 254.6 | 347.95 | 0 | 96.925 |
|
| Agilent44K |
N/A |
| Proteome |
| DBTMEE | Gene | Entrez | GV_seqCount | MII_seqCount | Zygote_seqCount | GV_specCount | MII_specCount | Zygote_specCount |
|---|
0003151
(v1.0) | Axin1 | 12005
| 0 | 0 | 1 | 0 | 0 | 2 |
|
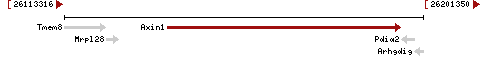
 57 Terms
57 Terms