DBTMEE:0004381
| Gene | Cdc45 |
| Synonyms | Cdc45l |
| Description | cell division cycle 45 |
| Locus | NM_001161623 UCSC: chr16:18780540-18811732(-) OLP: chr16:18780540-18811732(-)
NM_009862 UCSC: chr16:18780540-18812065(-) OLP: chr16:18780540-18812065(-)
|
| DB links |
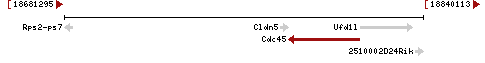
Entrez 12544
MGI 1338073
Ensembl ENSMUSG00000000028
Vega OTTMUSG00000026176
|
| Feature | protein-coding |
| GO_BP |
-
 4 Terms
|
| GO_MF | |
| GO_CC |
-
 2 Terms
|
| KEGG |
-
 1 Terms
|
| Ver2_FPKM |
see DBTMEE:2004270 (v2.0)
|
| SPR_Histone_Erkek_5KaroundTSS |
N/A |
| SPR_Histone_Erkek_2KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_5KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_2KaroundTSS |
N/A |
| SOLiD_OocyteTo4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | SPR_FPKM | Oo_FPKM | 1C_FPKM | 2C_FPKM | 4C_FPKM | MEF_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 35.3062 | 39.5386 | 92.7767 | 62.53 | 15.6373 | |
|
| SOLiD_Illumina_Oo_1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | 1C_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 36.9159 | 1.39103 | 43.1374 |
|
| SOLiD_Partheno1C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 1C_FPKM | p1C_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 36.0336 | 39.1416 | 38.8472 |
|
| SOLiD_Illumina_Oo_p1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | p1C_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 36.9374 | 1.37198 | 42.6063 |
|
| SOLiD_Partheno4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | 2C_FPKM | 4C_FPKM | p4C_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 85.8968 | 60.3753 | 57.5848 |
|
| Illumina_Oo_2C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 2C_FPKM |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 9.20275 | 57.0835 |
|
| SOLiD_SC_Oo_REPLICA_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_1_1 | Oo_2_1 |
|---|
0004381
(v1.0) | Cdc45 | NM_001161623,NM_009862 | 12544
| 53.9677 | 50.4759 |
|
| NIA60mer |
| DBTMEE | Array_Feature_Number | Oligo_Parent_Clone | Unigene | Gene | Entrez | U_Cluster | NAP_Cluster | Gene_U/NAP | TIGR | SwissProt | EST | Oo | 1C | 2C | 4C | 8C | Morula | Blastocyst |
|---|
0004381
(v1.0) | 259 | C0019C07-3 | Mm.1248 | Cdc45l | 12544
| U036865 | NAP099175-001 | Cdc45l; | TC875769 | Q9Z1X9 | U,F,2 | 3.9861 | 4.0839 | 3.8114 | 3.9756 | 3.3753 | 3.4268 | 3.257 |
|
| MOE430 |
| DBTMEE | Probe_ID | Unigene | Gene | RefSeq | Entrez | Oo | 1C | 2C | 8C | Blastocyst |
|---|
0004381
(v1.0) | 1416575_at | Mm.1248 | Cdc45 | NM_009862.2,NM_001161623.1 | 12544
| 254.975 | 484.325 | 567.475 | 395.75 | 249.475 |
|
| Agilent44K |
| DBTMEE | Probe_ID | Gene | Acc | Entrez | Oo1_xFold | Oo1_pVal | Oo1_Flag | Oo2_xFold | Oo2_pVal | Oo2_Flag |
|---|
0004381
(v1.0) | A_51_P401451 | Cdc45l | NM_009862 | 12544
| 1 | 1 | P | 1.235 | 0.372 | P |
|
| Proteome |
N/A |
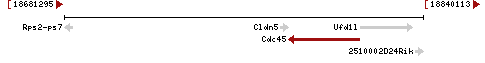
 4 Terms
4 Terms