DBTMEE:0009366
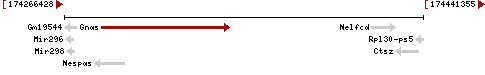
| Gene | Gnas |
| Synonyms | G alpha s
Galphas
Gnas1
Gnasxl
Gs alpha
Gs-alpha
Gsa
Nesp
Nesp55
Nespl
neuroendocrine-specific Golgi protein p55 isoform 1
Oedsml
P1
P2
P3
XLalphas |
| Description | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| Locus | NM_019690 UCSC: chr2:174109821-174163585(+) OLP: chr2:174109821-174163585(+)
NM_022000 UCSC: chr2:174109821-174172243(+) OLP: chr2:174109821-174172243(+)
NM_010309 UCSC: chr2:174123360-174172243(+) OLP: chr2:174123360-174172243(+)
NM_201617 UCSC: chr2:174123360-174126303(+) OLP: chr2:174123360-174126303(+)
NM_201618 UCSC: chr2:174123360-174172243(+) OLP: chr2:174123360-174172243(+)
NM_001077507 UCSC: chr2:174123360-174172243(+) OLP: chr2:174123360-174172243(+)
NR_003258 UCSC: chr2:174153415-174172243(+) OLP: chr2:174153415-174172243(+)
NM_201616 UCSC: chr2:174155590-174172243(+) OLP: chr2:174155590-174172243(+)
NM_001077510 UCSC: chr2:174155590-174172243(+) OLP: chr2:174155590-174172243(+)
|
| DB links |
Entrez 14683
MGI 95777
Ensembl ENSMUSG00000027523
Vega OTTMUSG00000016388
|
| Feature | protein-coding |
| GO_BP |
-
 30 Terms
|
| GO_MF |
-
 17 Terms
|
| GO_CC |
-
 18 Terms
|
| KEGG |
-
 16 Terms
|
| Ver2_FPKM |
see DBTMEE:2009244 (v2.0)
|
| SPR_Histone_Erkek_5KaroundTSS |
N/A |
| SPR_Histone_Erkek_2KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_5KaroundTSS |
N/A |
| SPR_Methy_Kobayashi_2KaroundTSS |
N/A |
| SOLiD_OocyteTo4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | SPR_FPKM | Oo_FPKM | 1C_FPKM | 2C_FPKM | 4C_FPKM | MEF_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 172.879 | 176.676 | 10.9996 | 43.2463 | 168.257 | |
|
| SOLiD_Illumina_Oo_1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | 1C_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 180.761 | 289.429 | 192.757 |
|
| SOLiD_Partheno1C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 1C_FPKM | p1C_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 176.44 | 174.902 | 211.99 |
|
| SOLiD_Illumina_Oo_p1C_Sperm_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | SPR_FPKM | p1C_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 180.866 | 285.465 | 232.503 |
|
| SOLiD_Partheno4C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | 2C_FPKM | 4C_FPKM | p4C_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 10.1839 | 41.756 | 55.6303 |
|
| Illumina_Oo_2C_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_FPKM | 2C_FPKM |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 209.795 | 34.6303 |
|
| SOLiD_SC_Oo_REPLICA_FPKM_gene_guided |
| DBTMEE | Gene | RefSeq | Entrez | Oo_1_1 | Oo_2_1 |
|---|
0009366
(v1.0) | Gnas | NM_001077507,NM_001077510,NM_019690,NM_022000,NM_201616,NM_201617,NM_201618 | 14683
| 38.868 | 35.0064 |
|
| NIA60mer |
| DBTMEE | Array_Feature_Number | Oligo_Parent_Clone | Unigene | Gene | Entrez | U_Cluster | NAP_Cluster | Gene_U/NAP | TIGR | SwissProt | EST | Oo | 1C | 2C | 4C | 8C | Morula | Blastocyst |
|---|
0009366
(v1.0) | 12712 | H3150F07-5 | | Gnas | 14683
| U002926 | NAP093468-001 | Gnas;Nespl; | TC869471,TC869469,TC869468,TC819197,TC819195,TC819194 | Q9Z1N8 | U,F,2 | 2.7591 | 2.9842 | 2.8442 | 2.5767 | 3.0124 | 2.6976 | 2.8886 |
|
| MOE430 |
| DBTMEE | Probe_ID | Unigene | Gene | RefSeq | Entrez | Oo | 1C | 2C | 8C | Blastocyst |
|---|
0009366
(v1.0) | 1450186_s_at | Mm.125770 | Gnas | NM_201618.1,NM_010309.3,NM_001077507.1,NM_201617.1,NM_022000.2,NM_201616.1,NM_001077510.2,NM_019690.2 | 14683
| 421.1 | 479.85 | 293.55 | 178.125 | 239.7 |
0009366
(v1.0) | 1453413_at | Mm.125770 | Gnas | NM_201618.1,NM_010309.3,NM_001077507.1,NM_201617.1,NM_022000.2,NM_201616.1,NM_001077510.2,NM_019690.2 | 14683
| 38.45 | 0 | 0 | 57.35 | 32.15 |
|
| Agilent44K |
| DBTMEE | Probe_ID | Gene | Acc | Entrez | Oo1_xFold | Oo1_pVal | Oo1_Flag | Oo2_xFold | Oo2_pVal | Oo2_Flag |
|---|
0009366
(v1.0) | A_52_P112182 | Gnas | S49980 | 14683
| 1 | 1 | P | 0.861 | 0.565 | P |
0009366
(v1.0) | A_52_P112188 | Gnas | NM_010309 | 14683
| 1 | 1 | P | 0.733 | 0.383 | P |
|
| Proteome |
| DBTMEE | Gene | Entrez | GV_seqCount | MII_seqCount | Zygote_seqCount | GV_specCount | MII_specCount | Zygote_specCount |
|---|
0009366
(v1.0) | Gnas | 14683
| 6 | 3 | 3 | 8 | 3 | 3 |
|

 30 Terms
30 Terms