Tutorials: Learn How to Use DBTMEE
CASE 1: Identification of Fertilization-specific Genes (Not Expressed in Parthenogenetic Development)
CASE 2: Inference of the TFs that Potentially Bind to Genes Expressed after Fertilization
CASE 3: Inferring the Effect of Histone Modifications in Sperm on Early Embryo Development
CASE 4: Identification of Potential Factors that Enhance Reprogramming to Induced Pluripotent Stem Cells
CASE 1: Identification of Fertilization-specific Genes (Not Expressed in Parthenogenetic Development)
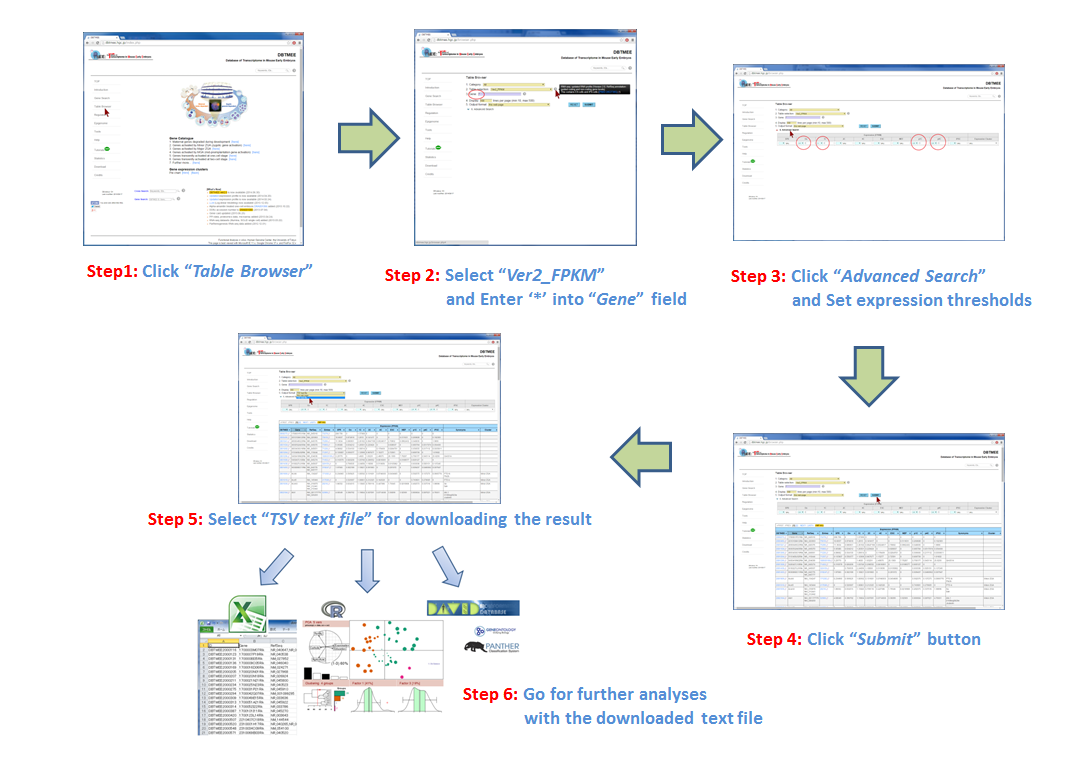
Question: Which genes are not expressed at both oocytes and parthenotes, but expressed immediately after fertilization? (Is it sperm-effect?) "Advanced Search" option in the Table Browser provides an 'AND' search operator by filtering gene expression levels (FPKMs) at stages and cells. For retrieving genes that are not expressed at parthenotes, this option must be set with specific FPKM values for p1C (parthenogenetic one cell stage) and/or p4C (parthenogenetic four cell stage). In this tutorial, after entering an asterisk (*, a wildcard that matches all gene names) into the Gene field (Step 2), we have set thresholds "<= 1.0" for p1C and p4C (Step 3). This set removes genes that have over 1.0 FPKMs at parthenotes, and returns genes that have any FPKM values at oocyte and/or normal development. For identifying fertilization-specific genes that are not expressed at oocytes but expressed after fertilization, we have additionally set "<=1.0" for Oo (Oocyte) and >1.0 for 1C (one-cell stage) (Step 3). The query result can be downloaded in the tab-delimited text file (Step 5). Then, users can further analyze the result by other software and tools, such as Excel, R, Gene Ontology, etc (Step 6). |
[Step 1]
 [Step 2] 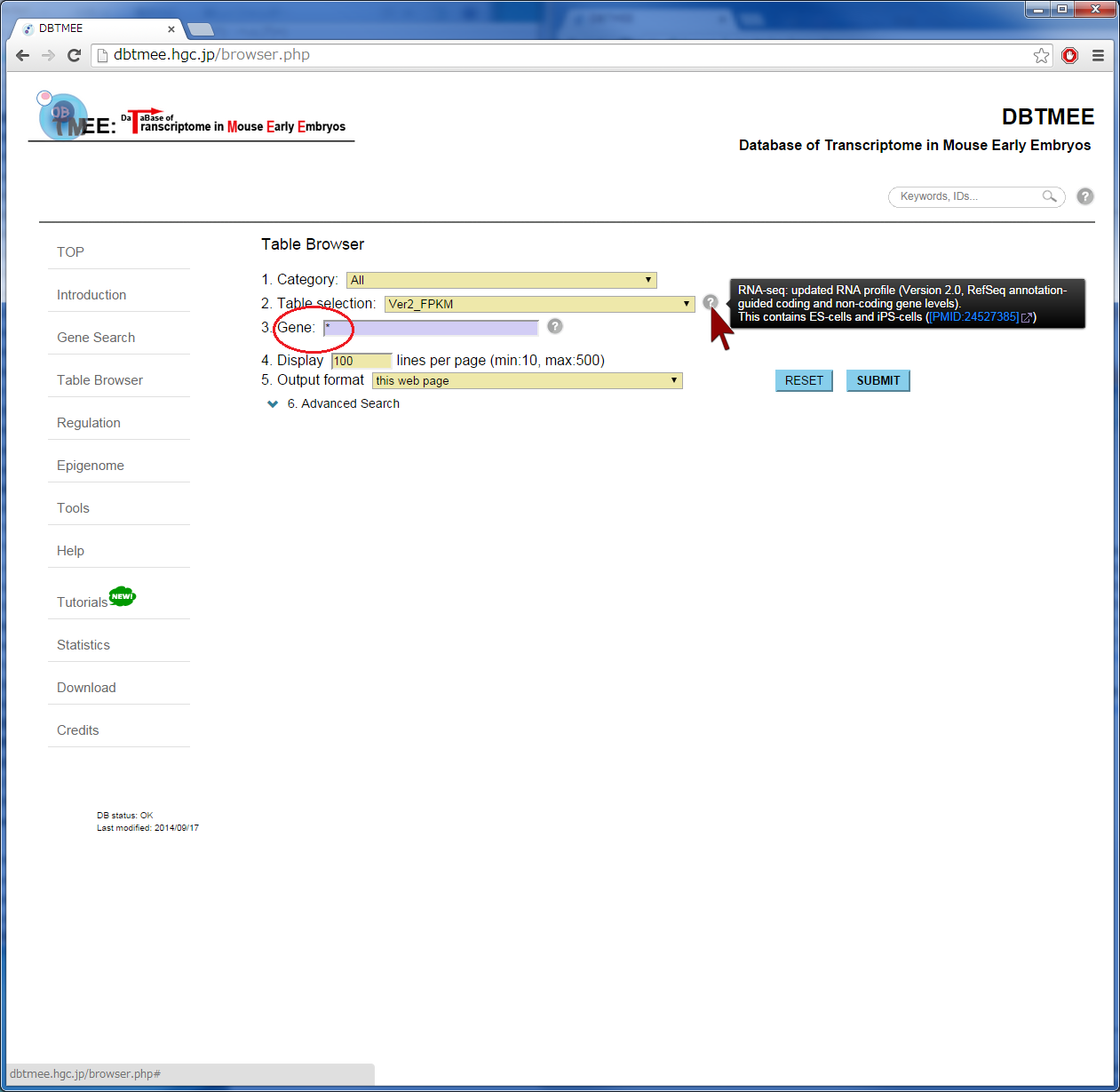 [Step 3]  [Step 4]  [Step 5] 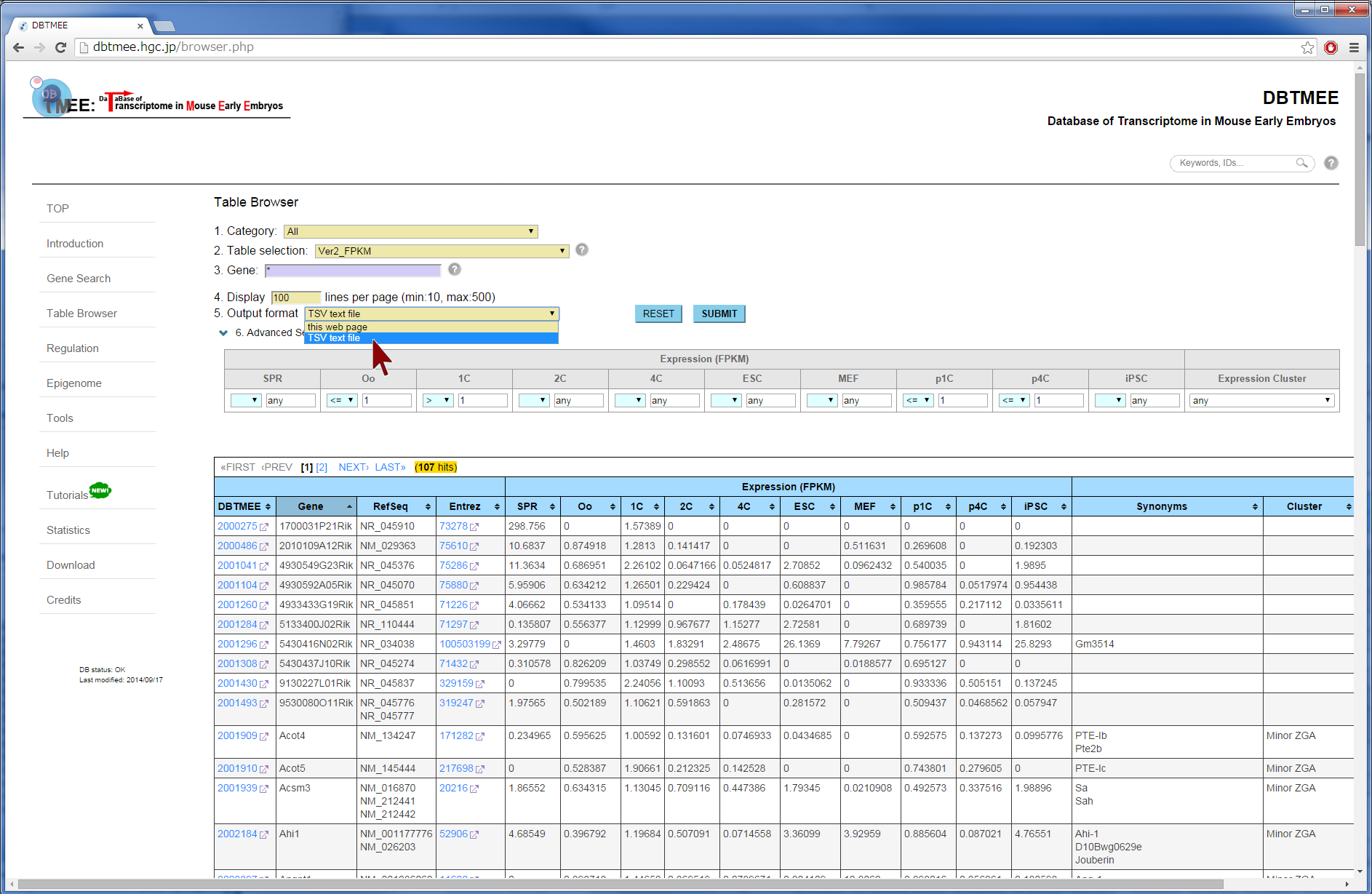 [Step 6] 
|
