Tutorials: Learn How to Use DBTMEE
CASE 1: Identification of Fertilization-specific Genes (Not Expressed in Parthenogenetic Development)
CASE 2: Inference of the TFs that Potentially Bind to Genes Expressed after Fertilization
CASE 3: Inferring the Effect of Histone Modifications in Sperm on Early Embryo Development
CASE 4: Identification of Potential Factors that Enhance Reprogramming to Induced Pluripotent Stem Cells
CASE 4: Identification of Potential Factors that Enhance Reprogramming to Induced Pluripotent Stem Cells (iPSCs)
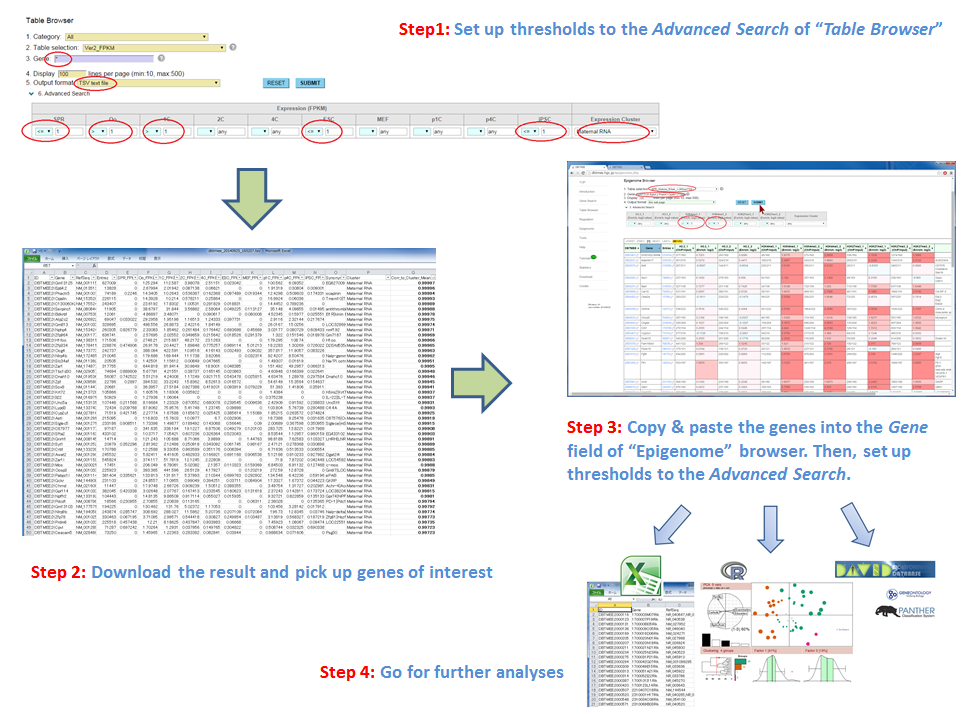
It has been reported that the factors enriched in oocytes and zygotes are good candidates to enhance the reprogramming of somatic cells to iPSCs; e.g. Glis1 (Maekawa et al. PUBMED), Hist1h2aa (TH2A), Hist1h2ba (TH2B) (Shinagawa et al. PUBMED). The characteristics of these factors are high gene expression levels at oocytes and zygotes (Maternal RNA category). In contrast, they exhibit low gene expression levels at ESCs and iPSCs [Step0-1, 0-2]. Moreover, though these TFs are the genes which are not expressed in sperm, they are significantly enriched by H3K4me3 (an active histone mark) in sperm [Step0-3, 0-4, 0-5]. H1foo exhibits the similar expression pattern, but H1foo is marked by neither histones in sperm. This linker histone does not enhance the generation of iPSCs (Shinagawa et al, PUBMED). To identify such potential factors for the reprogramming, we set up thresholds to the Advanced Search option in Table Browser [Step 1];
<=1 for SPR, >1 for Oo, >1 for 1C, <=1 for ESC, <=1 for iPSC, "Maternal RNA" for Expression Cluster.
Although this set returns 365 genes, we picked up 182 genes that are >0.98 in the "Corr_to_Cluster_Mean" column (correlation to the mean of Maternal RNA category). Then, we pasted the gene names into the Gene field of the Epigenome Browser, and set up thresholds;
>1 for H3K4me3_1, >1 for H3K4me3_2.
This set returns 60 genes including six TFs (Glis1, Lhx8, Zfp78, Nod2, Prdm6, Sox8) [Step 3];
9330182L06Rik Adamts17 Astl Asz1 Bai3 Car10 Cela2a Chrm4 Cmya5 Cnga4 Crb1 Cyp2u1 Dnah10 Fam189a1 Fbxw18 Fgf8 Flt1 Gckr Glis1 Gpr137c Grid1 Hist1h2aa Hist1h2ba Il22 Kctd7 Lhx8 Lypd3 Mast4 Mei4 Mfsd2a Nod2 Nphp4 Nrg3 Obscn Oog4 Oprd1 Ppp1r3d Prdm6 Pthlh Ptprm Rad51ap2 Rbm20 Reln Rimkla Sema6d Slc2a13 Slc9a4 Sox8 Syt2 Tac1 Tbc1d30 Tdrd1 Tmc3 Wnt7a Zar1 Zbtbd6 Zfp583 Zfp612 Zfp618 Zfp78.
This result might be helpful to identify genes that enhance the reprogramming of somatic cells to iPSCs. |
[Step 0-1]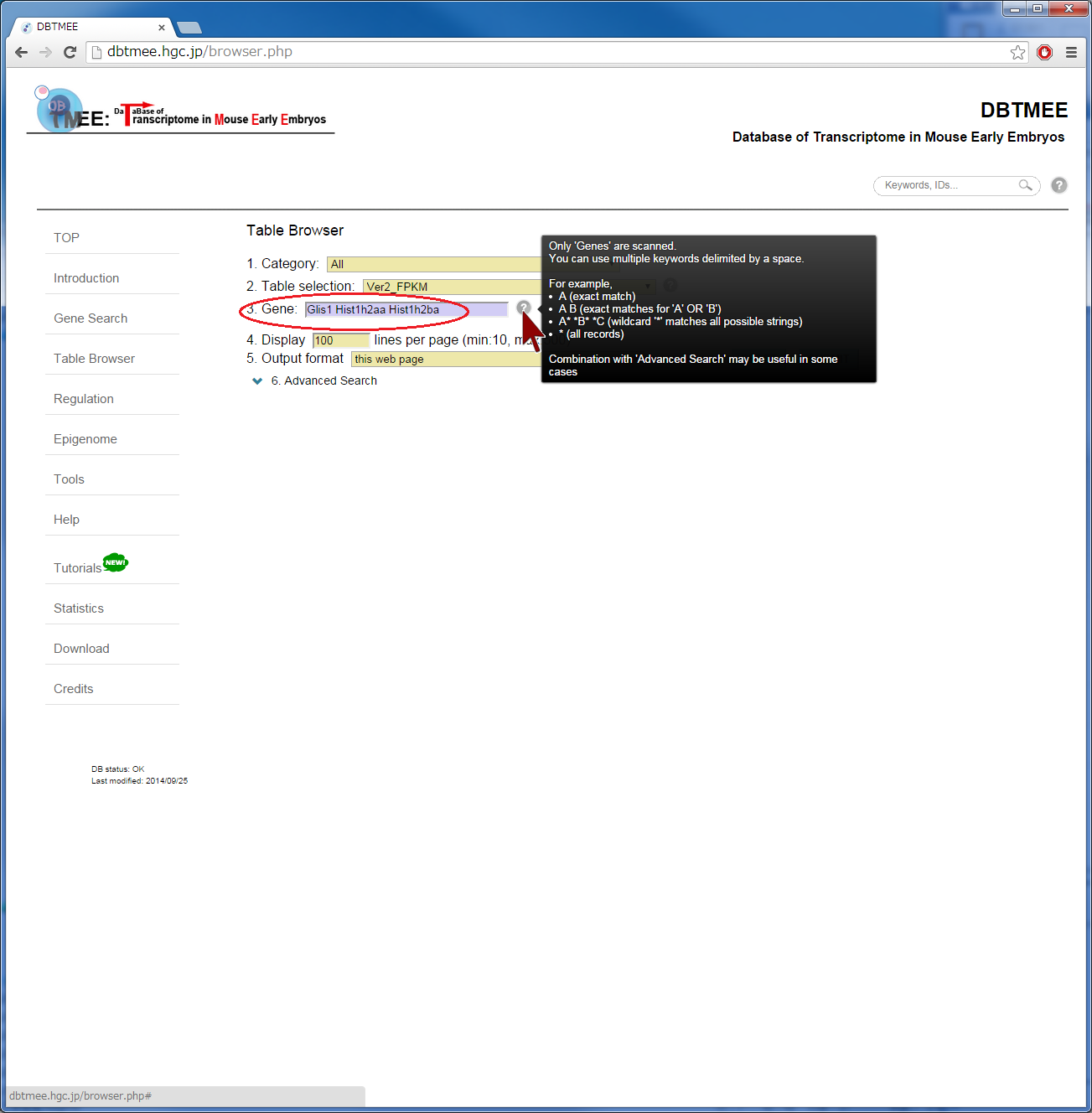 [Step 0-2]  [Step 0-3] 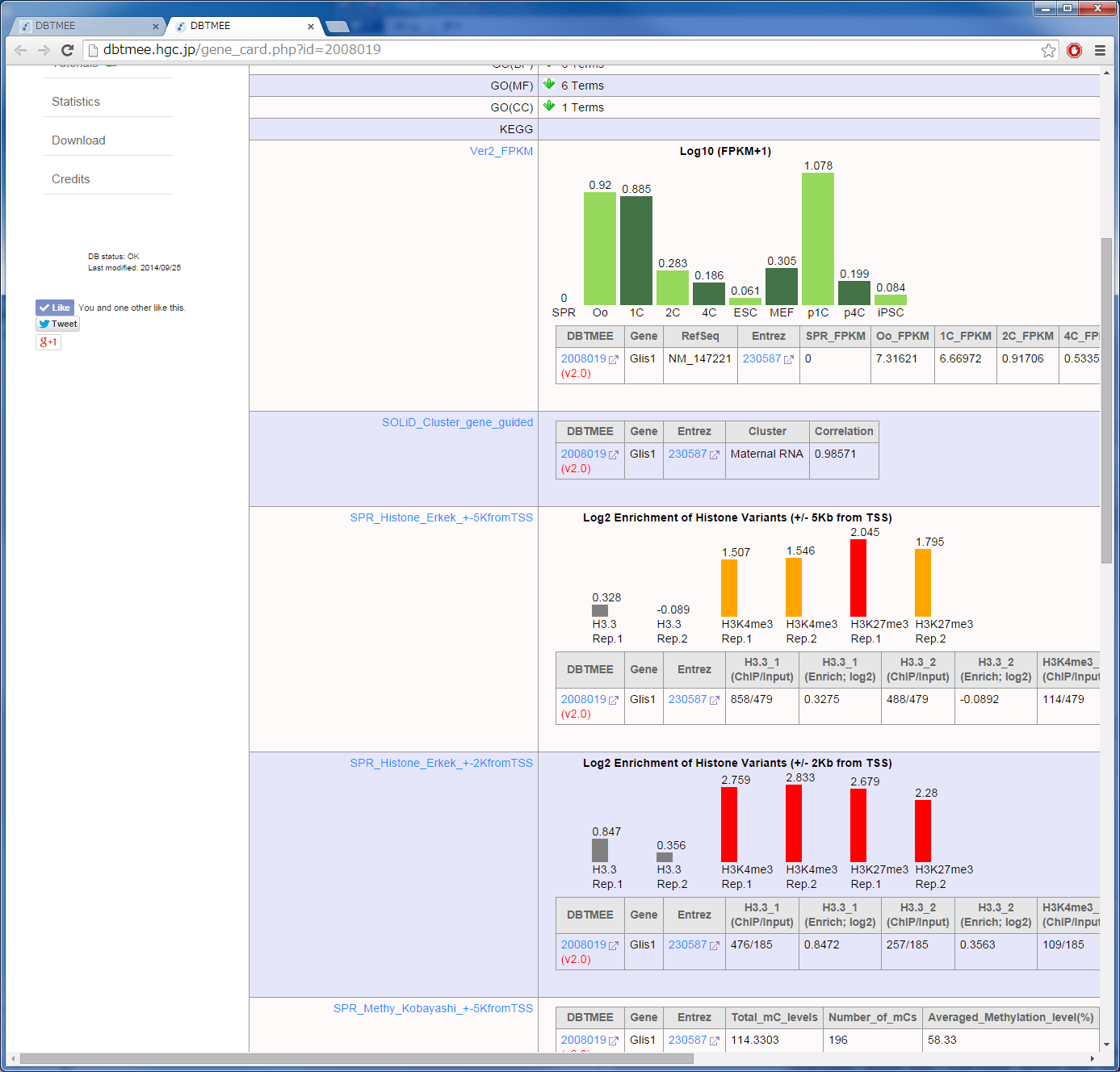 [Step 0-4] 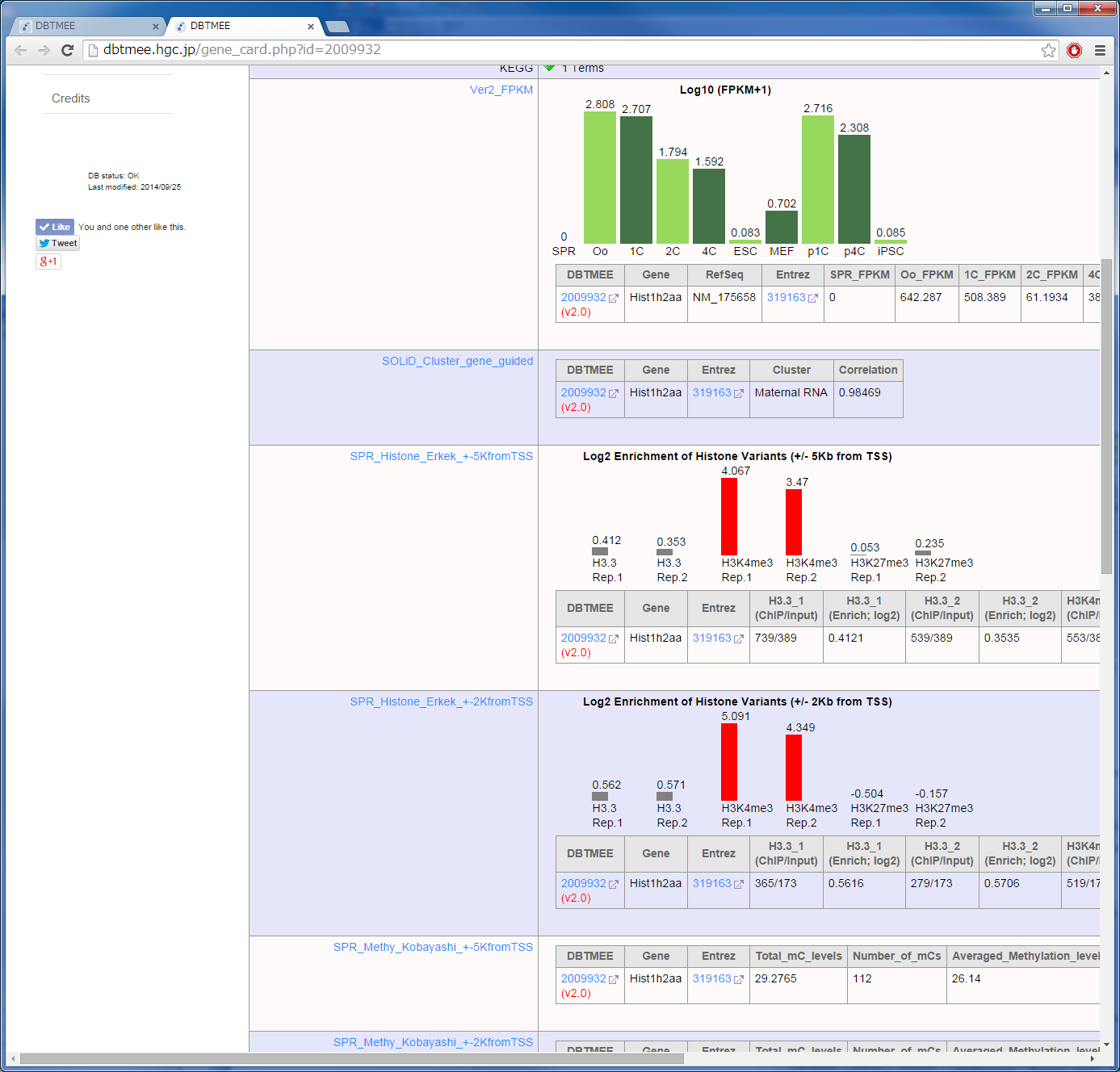 [Step 0-5] 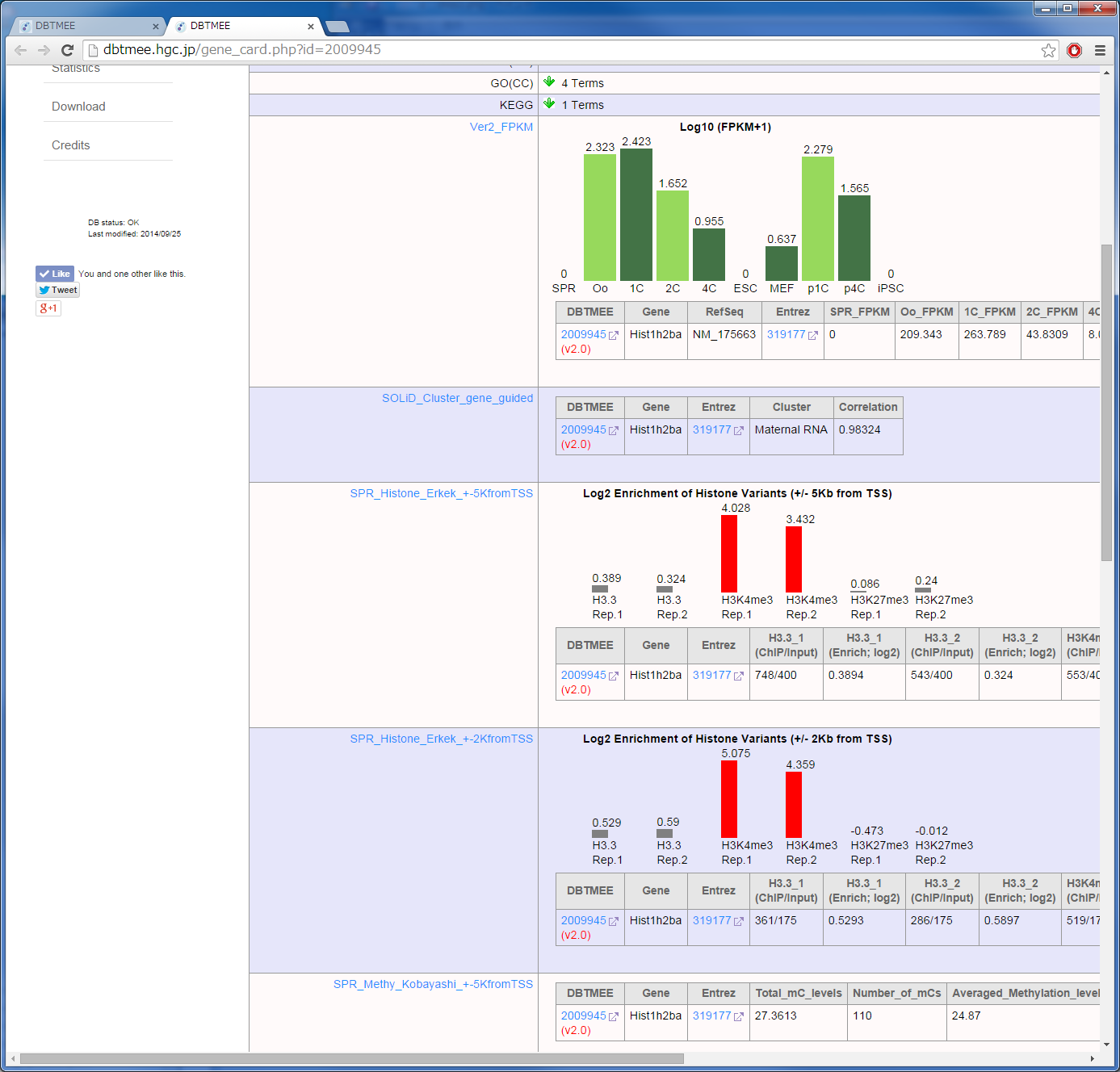 [Step 1] 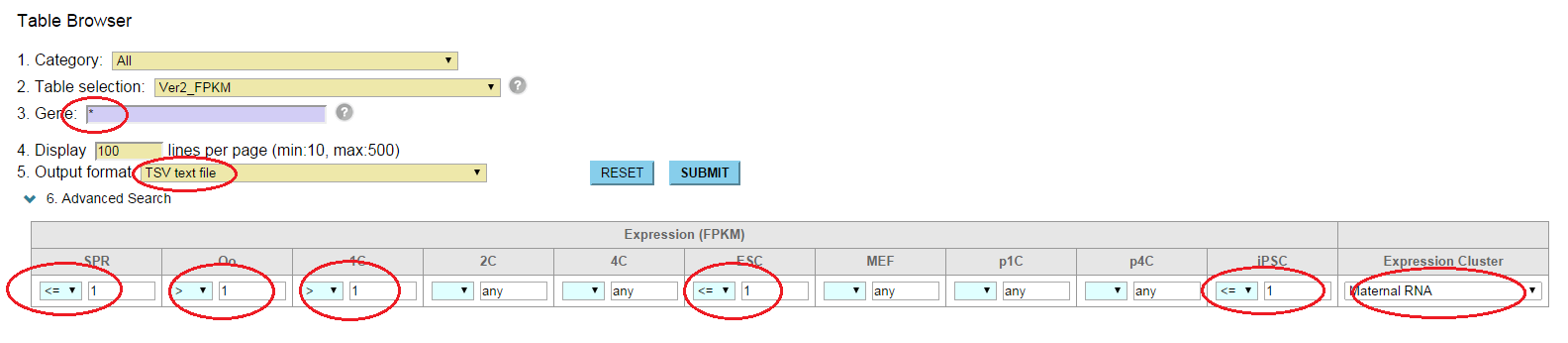 [Step 3]  [Step 4]  |
